Search
Asgard (archaea)

Asgard or Asgardarchaeota is a proposed superphylum consisting of a group of archaea that contain eukaryotic signature proteins. It appears that the eukaryotes, the domain that contains the animals, plants, and fungi, emerged within the Asgard, in a branch containing the Heimdallarchaeota. This supports the two-domain system of classification over the three-domain system.
Discovery and nomenclature
In the summer of 2010, sediments were analysed from a gravity core taken in the rift valley on the Knipovich ridge in the Arctic Ocean, near the Loki's Castle hydrothermal vent site. Specific sediment horizons previously shown to contain high abundances of novel archaeal lineages were subjected to metagenomic analysis. In 2015, an Uppsala University-led team proposed the Lokiarchaeota phylum based on phylogenetic analyses using a set of highly conserved protein-coding genes. The group was named for the shape-shifting Norse god Loki, in an allusion to the hydrothermal vent complex from which the first genome sample originated. The Loki of mythology has been described as "a staggeringly complex, confusing, and ambivalent figure who has been the catalyst of countless unresolved scholarly controversies", analogous to the role of Lokiarchaeota in the debates about the origin of eukaryotes.
In 2016, a University of Texas-led team discovered Thorarchaeota from samples taken from the White Oak River in North Carolina, named in reference to Thor, another Norse god. Samples from Loki's Castle, Yellowstone National Park, Aarhus Bay, an aquifer near the Colorado River, New Zealand's Radiata Pool, hydrothermal vents near Taketomi Island, Japan, and the White Oak River estuary in the United States contained Odinarchaeota and Heimdallarchaeota; following the Norse deity naming convention, these groups were named for Odin and Heimdall respectively. Researchers therefore named the superphylum containing these microbes "Asgard", after the home of the gods in Norse mythology. Two Lokiarchaeota specimens have been cultured, enabling a detailed insight into their morphology.
Description
Proteins
Asgard members encode many eukaryotic signature proteins, including novel GTPases, membrane-remodelling proteins like ESCRT and SNF7, a ubiquitin modifier system, and N-glycosylation pathway homologs.
Asgard archaeons have a regulated actin cytoskeleton, and the profilins and gelsolins they use can interact with eukaryotic actins. In addition, Asgard archaea tubulin from hydrothermal-living Odinarchaeota (OdinTubulin) was identified as a genuine tubulin. OdinTubulin forms protomers and protofilaments most similar to eukaryotic microtubules, yet assembles into ring systems more similar to FtsZ, indicating that OdinTubulin may represent an evolution intermediate between FtsZ and microtubule-forming tubulins. They also seem to form vesicles under cryogenic electron microscopy. Some may have a PKD domain S-layer. They also share the three-way ES39 expansion in LSU rRNA with eukaryotes. Gene clusters or operons encoding ribosomal proteins are often less conserved in their organization in the Asgard group than in other Archaea, suggesting that the order of ribosomal protein coding genes may follow the phylogeny.
Metabolism
Asgard archaea are generally obligate anaerobes, though Kariarchaeota, Gerdarchaeota and Hodarchaeota may be facultative aerobes. They have a Wood–Ljungdahl pathway and perform glycolysis. Members can be autotrophs, heterotrophs, or phototrophs using heliorhodopsin. One member, Candidatus Prometheoarchaeum syntrophicum, is syntrophic with a sulfur-reducing proteobacteria and a methanogenic archaea.
The RuBisCO they have is not carbon-fixing, but likely used for nucleoside salvaging.
Ecology
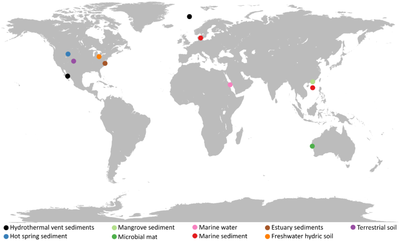
Asgard are widely distributed around the world, both geographically and by habitat. Many of the known clades are restricted to sediments, whereas Lokiarchaeota, Thorarchaeota and another clade occupy many different habitats. Salinity and depth are important ecological drivers for most Asgard archaea. Other habitats include the bodies of animals, the rhizosphere of plants, non-saline sediments and soils, the sea surface, and freshwater. In addition, Asgard are associated with several other microorganisms.
Eukaryote-like features in subdivisions
The phylum Heimdallarchaeota was found in 2017 to have N-terminal core histone tails, a feature previously thought to be exclusively eukaryotic. Two other archaeal phyla, both outside of Asgard, were found to also have tails in 2018.
In January 2020, scientists found Candidatus Prometheoarchaeum syntrophicum, a member of the Lokiarcheota, engaging in cross-feeding with two bacterial species. Drawing an analogy to symbiogenesis, they consider this relationship a possible link between the simple prokaryotic microorganisms and the complex eukaryotic microorganisms occurring approximately two billion years ago.
Phylogeny
The phylogenetic relationships of the Asgard archaea have been studied by several teams in the 21st century. Varying results have been obtained, for instance using 53 marker proteins from the Genome Taxonomy Database. In 2023, Eme, Tamarit, Caceres and colleagues reported that the Eukaryota are deep within Asgard, as sister of Hodarchaeales within the Heimdallarchaeia.
Taxonomy
In the depicted scenario, the Eukaryota are deep in the tree of Asgard. A favored scenario is syntrophy, where one organism depends on the feeding of the other. An α-proteobacterium was incorporated to become the mitochondrion. In culture, extant Asgard archaea form various syntrophic dependencies. Gregory Fournier and Anthony Poole have proposed that Asgard is part of "the Eukaryote tree", forming a superphylum they call "Eukaryomorpha" defined by "shared derived characters" (eukaryote signature proteins).
The taxonomy is uncertain and the phylum names are therefore somewhat speculative. The list of phyla is based on the List of Prokaryotic names with Standing in Nomenclature (LPSN) and National Center for Biotechnology Information (NCBI).
- Phylum Baldrarchaeota Caceres 2019
- Phylum Borrarchaeota Liu et al. 2021
- Phylum Freyrarchaeota corrig. Caceres 2019
- Phylum Friggarchaeota Caceres 2019
- Phylum Gefionarchaeota Caceres 2019
- Phylum Gerdarchaeota Cai et al. 2020
- Phylum Heimdallarchaeota Zaremba-Niedzwiedzka et al. 2017
- Phylum Helarchaeota Seitz et al. 2019
- Phylum Hermodarchaeota Liu et al. 2021
- Phylum Hodarchaeota Liu et al. 2021
- Phylum Idunnarchaeota Caceres 2019
- Phylum Kariarchaeota Liu et al. 2021
- Phylum Lokiarchaeota Spang et al. 2015
- Phylum Njordarchaeota Xie et al. 2022
- Phylum Odinarchaeota Zaremba-Niedzwiedzka et al. 2017
- Phylum Sifarchaeota Farag et al. 2020
- Phylum Sigynarchaeota Xie et al. 2022
- Phylum Thorarchaeota Baker 2015
- Phylum Tyrarchaeota Xie et al. 2022
- Phylum Wukongarchaeota Liu et al. 2021
Genomic elements
Viruses
Several family-level groups of viruses associated with Asgard archaea have been discovered using metagenomics. The viruses were assigned to Lokiarchaeia, Thorarchaeia, Odinarchaeia and Helarchaeia hosts using CRISPR spacer matching to the corresponding protospacers within the viral genomes. Two groups of viruses (called 'verdandiviruses') are related to archaeal and bacterial viruses of the class Caudoviricetes, i.e., viruses with icosahedral capsids and helical tails; two other distinct groups (called 'skuldviruses') are distantly related to tailless archaeal and bacterial viruses with icosahedral capsids of the realm Varidnaviria; and the third group of viruses (called wyrdviruses) is related to archaea-specific viruses with lemon-shaped virus particles (family Halspiviridae). The viruses have been identified in deep-sea sediments and a terrestrial hot spring of the Yellowstone National Park. All these viruses display very low sequence similarity to other known viruses but are generally related to the previously described prokaryotic viruses, with no meaningful affinity to viruses of eukaryotes.
Mobile genetic elements
In addition to viruses, several groups of cryptic mobile genetic elements have been discovered through CRISPR spacer matching to be associated with Asgard archaea of the Lokiarchaeia, Thorarchaeia and Heimdallarchaeia lineages. These mobile elements do not encode recognizable viral hallmark proteins and could represent either novel types of viruses or plasmids.
See also
- List of Archaea genera
References
External links
- Traci Watson: The trickster microbes that are shaking up the tree of life, in: Nature, 14 May 2019
Text submitted to CC-BY-SA license. Source: Asgard (archaea) by Wikipedia (Historical)
Owlapps.net - since 2012 - Les chouettes applications du hibou